The SNIC Science Cloud is a project run by the Swedish National Infrastructure for Computing. The goal is to investigate if and how cloud resources should be provided as a complement to the more traditional HPC-resources. An important part of this investigation is our pilot users. The project has run in two main phases:
1. 2013-2014: A small-scale pilot system and a small number (3-5) of predefined pilot use-cases, chosen to highlight the utility of cloud resources. Advanced user support in the form of SNIC Application Experts (AEs) were available to assist the pilot users.
2. 2015-2016: A scaled up infrastructure with more resources, and an open process for user-initiated project requests. In this phase, we have concentrated on training workshops and creating open source tutorials instead of targeted advanced support.
In the summer 2016 we reported on the growth of project requests and promised to follow up with a more in-depth analysis of who these users are and what they are doing. We have looked in our project database at all the projects that registered for SSC resources in 2015-2016 (as of Oct. 11). During the period, we have had 57 project requests.
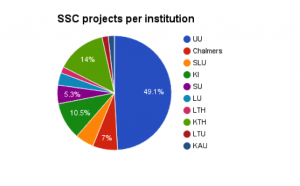
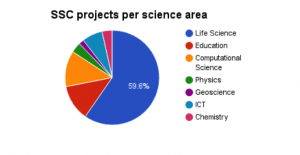
As can be seen, Life Science users dominate. Of these, a large fraction are affiliated with SciLifeLab/NBIS. This is expected since the Bioinformatics community was an early adopter of service-oriented computing, and since their applications often have the need to integrate multiple software. Many Swedish universities are represented, but Uppsala University (UU), The Royal Institute of Technology (KTH) and the Karolinska Institute (KI) dominate. This is likely a consequence of the fact that these institutions’ involvement in SciLifeLab, and that both KTH and UU has served as hosts for the SSC project, presumably increasing awareness of SSC amongst the scientists.
In the period Sep 11-Oct 11, a total of 47000 instance hours were deployed in 31 different projects. Using a reference instance type (flavor) with 4 VCPUs and 8GB RAM, this corresponds to an average of 130 instances continuously deployed during the month. We will follow up on the resource usage patterns over time in future posts, when we have more fine-grained data.
So what are the users doing with the SSC OpenStack resources? It appears that development and testing of software and services, as well as exploring the cloud computing paradigm for old and new types of applications are still the dominating use case. In common to most projects is the need for flexible customization of the computing environment, made possible by virtualization. Many projects also want to provide their solutions as services to serve their own specific community.
Some projects are making more substantial use of the IaaS resources, making use of advanced tools for contextualization, automation and orchestration to achieve quite a diverse range of objectives. In common to all these projects is that they have access to own expertise on distributed and cloud computing in the project groups. To serve as an inspiration to new users, we have during 2016 highlighted some of them as user success stories:
Elastic proteomics analysis in the Malmstroem Lab.
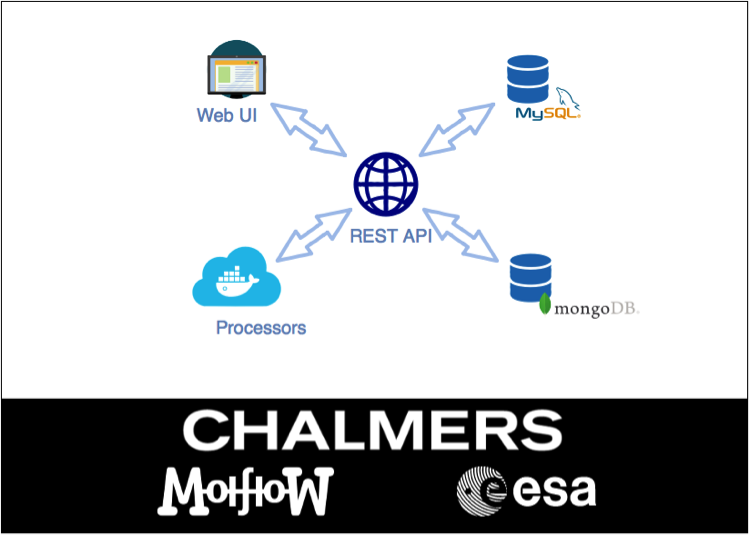
Processing ozone data from the Odin satellite at Chalmers University of Technology.
Estimation of failure probabilities with applications in underground porous media flows.
Virtual Research Environment for Clinical Metabolomics.
So what will happen in 2017? A projection is hard to provide, but given the global trend that private/community IaaS becomes more and more common also in academia, observations made by our partners in the Nordic Glenna project, and with the momentum created via the European OpenScienceCloud initiative, we believe that the interest in cloud resources will keep increasing rapidly.
Fortunately, in the SSC project we are in a good position to meet an increased demand due to our architectural design based on regions, in which we can leverage previous generation HPC hardware at multiple geographic locations to quickly add compute hosts at low cost. We have now integrated resources at three HPC-centra, UPPMAX, C3SE and HPC2N and can if needed scale resources to over 5000 physical cores and 1PB of storage during 2017. This model also opens up for substantial user communities to enter SSC with their own dedicated regions. We also hope to start looking into public-private partnerships to secure a larger variety of SLA-backed resources and to allow for users to burst outside of the allocated quotas.